
This program uses eight python files and one input file as shown below.
1. binomial_v016.py (main program)
This is a main program. This contains reading process of a input file and
a main loop for calculation.
2. element_35.py
The element class in this file defines elements in the input files. Elements
are molecules or various cells or living individuals, and/or etc.
3. reaction_63.py
Classes in this file define several reactions for the elements in the input file.
4. utility_55.py
This program contains utilities for plotting process, file saving, time managing, and so on.
5. polymer_12.py
If the input file is about “polymer”, this file is used.This file supports polymer reaction process.
6. reactionFunctions.py
This file provides reaction process management codes.
7. setting_30.py
This file provides input file setting process management codes.
8. binomial_parameters_02.json
The program is developed by Spyder IDE 5.3.3 on Ubuntu 22.04.1 LT, 3 April 2023.
Examples
Here shows three examples of input files. Detailed expression are mentioned in the
README_v016.txt,
which is downloadable from this site. There are many other examples in it’s downloaded zip file.
1. inp_exponential_401.txt
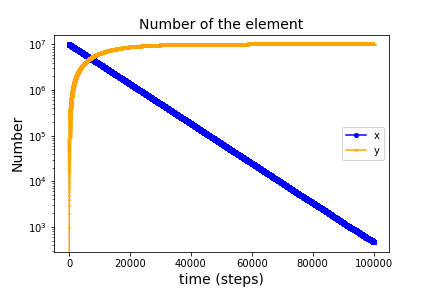
Calculation of this file shows an example of exponential decay.
The input file above needs items: *Time, *Element, *Reaction, and *Plot as follows.
The x becomes the y according to the reaction r1_1_100 (named by reaction type and name).
Element’s number is natural number more than zero. Reaction orders also are natural number.
** means comment.
*Time,
** start, end, console out, plot out, csv out, unit
0, 100000, 10000, 10000, 10000, steps
*Element
** 0:element name, 1:initial number, 2:plot color, 3:maeker
x, 10000000, Blue,
y, 0, Orange,
*Reaction, 1000000
** 0:reaction type, 1:name, 2:order, 3:element, 4:probability, 5:order, 6:element
r1_1, 100, 1, x, 0.0001, 1, y
*Plot, log
** second item is “linear” or “log”.
x, y
2. inp_SIR_010.txt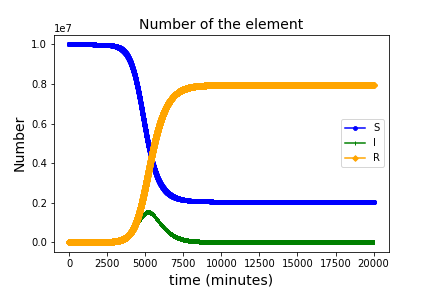
You can define SIR model as follows.
S is susceptible, I is infected, and R is recovered. Three types of populations are defined in *Element. Two reactions, r2_1_100 and r1_1_200 are defined in *Reaction.
This algorithm easily calculates those types of reactions.
*Time,
** start, end, console out, plot out, csv out, unit
0, 20000, 5000, 20000, 2000, minutes
*Element
** 0:element name, 1:initial number, 2:plot color, 3:marker
S, 10000000, Blue
I, 20, Green
R, 0, Orange
*Reaction, 10000000
r2_1, 100, 1, S, 1, I, 0.005, 2, I
r1_1, 200, 1, I, 0.0025, 1, R
*Plot, linear
** second items are ‘linear’ or ‘log’.
S, I, R
3. inp_feedback_408.txt
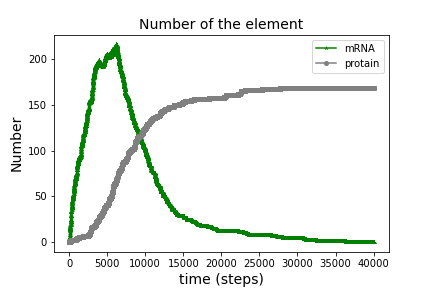
In a biological cell, feedback loop of gene system is primitive and essential reaction system.
This input file shows one DNA’s feedback loop reaction. RNA and amino-acid are collapsed into one type of them for simplicity. Many types of reactions are used.
*Time,
** start, end, console out, plot out, csv out, unit
0, 50000, 10000, 10000, 10000, steps
*Element
** 0:element name, 1:initial number, 2:plot color, 3:real name, 4:type, 5:volume , 6: volume unit
rna, 100000,
mRNA, 0, Green
amino, 100000,
protain, 0, Gray
s, 100, Orange
DNA, 1, blue
DNA_d, 0, Red
Ribo, 100,
*Reaction, 1000000
r3_3, 100, 1, DNA, 1, s, 10, rna, 1000, 1, DNA, 1, s, 1, mRNA
r3_3, 200, 1, mRNA, 1, Ribo, 10, amino, 10, 1, mRNA, 1, Ribo, 1, protain
r1_0, 300, 1, s, 0.0001
r1_0, 400, 1, mRNA, 0.0002
r2_1, 600, 1, DNA, 1, protain, 20, 1, DNA_d
r3_3, 700, 1, DNA_d, 1, s, 10, rna, 0.001, 1, DNA_d, 1, s, 1, mRNA
*Plot, linear
** second item is “linear” or “log”.
rna, amino
mRNA, protain
DNA, DNA_d
s,
Executing a program
Two excuting processes are abailable.
You prepare two holders in a master holder at first. One holder contains the six program files.
The other holder should contains a input text file like as mentioned above.
(Look at “Folder structure_v016.pdf”)
1. Spyder
Open Spyder in anaconda3. Open the main file, binomial_v016.py from Spyder.
You should write input file name in the appropriate line as follows,
fName = ‘inp_exponentiial_401.txt’
“Run” command in Spyder executes the input file. Results files
will be put in a new holder which is created in the input file holder.
Figers written in *Plot of the input file are displayed in the Plot window of Spyder.
2. command line
Preparing holders are same as for 1.Spyder case.
Open a terminal, and change directory to the program-file located directory.
you write a command line in a terminal as follows.
$ python binomial_v016.py inp_exponential_401.txt
Enter the line and the program starts.
Result files will be located in a new created holder.
